Annotations
Click the Annotations tab after clicking the icon to access the Annotations window (Fig.). The Annotations window shows the list of the annotation files, which are utilized in the functional annotations applications, such as
-
-
- Gene Models (GFF/GTF);
- Pathway (GMT);
- Gene Ontology (GO);
- Antibiotic Resistance (ABR); and
- Variant Effect Predictor (VEP).
- Variations
-
The paginated window displays 10 annotations, by default, per page and can be increased to show 20, 40, 60, 80, 100 annotations. The search box (Fig.) on the top right corner of the page allows users to filter annotations with file names, type, organism name, or version. During upload, the annotation files undergo a rigorous validation process for file format, integrity, and content.
Examples:
-
-
- Gene Models should be compatible with the reference genome version.
- Pathway and Gene Ontology files are validated against the corresponding Gene Models.
-
Validation failed annotations are greyed out and displayed with a “Validation Failed” message. The failed annotations are not available for selection and should be deleted from the platform by the user.
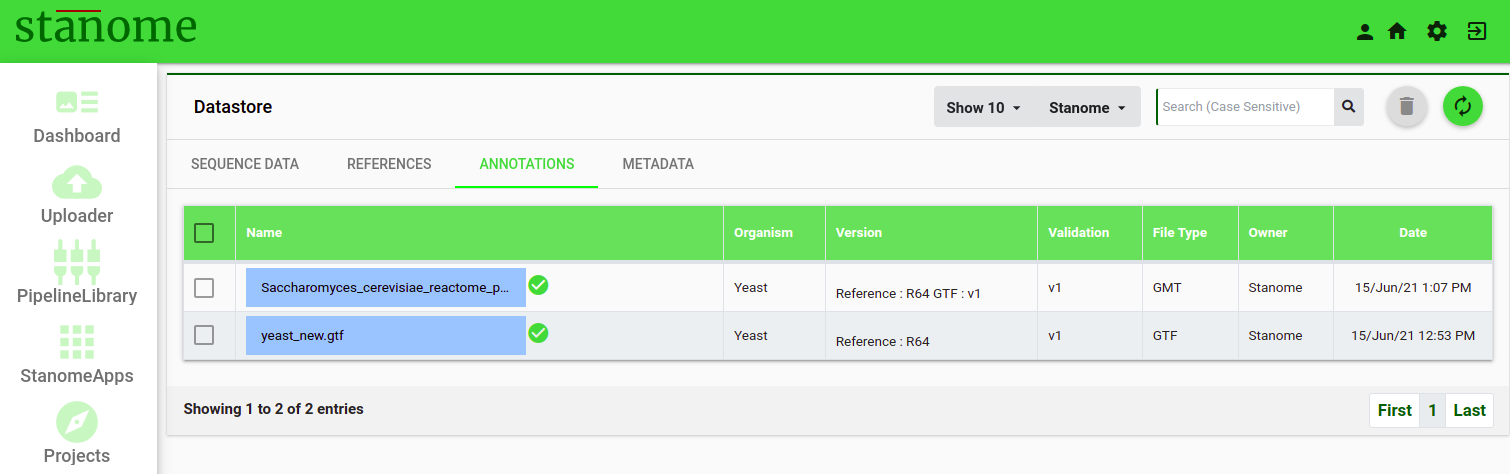
The list view shows seven fields :
- Name: Filename
- Organism: Organism to which annotation file corresponds to
- Version: Reference version to which the annotation file is associated
- Validation: Version of the annotation file
- FileType: Type of the file (Example: Pathway, GO, or VEP)
- Owner: Owner of the file
- Date: Displays the file upload date
